Introduction
nf-core/coproid is a bioinformatics pipeline that helps you identify the “true maker” of Illumina sequenced (Paleo)faeces by checking the microbiome composition and the endogenous host DNA.
It combines the analysis of putative host (ancient) DNA with a machine learning prediction of the faeces source based on microbiome taxonomic composition:
A. First coproID performs comparative mapping of all reads agains two (or three) target genomes (genome1, genome2, and potentially genome3) and computes a host-DNA species ratio (NormalisedProportion). B. Then coproID performs metagenomic taxonomic profiling, and compares the obtained profiles to modern reference samples of the target species metagenomes. Using machine learning, coproID then estimates the host source from the metagenomic taxonomic composition (SourcepredictProportion). C. Finally, coproID combines the A and B proportions to predict the likely host of the metagenomic sample.
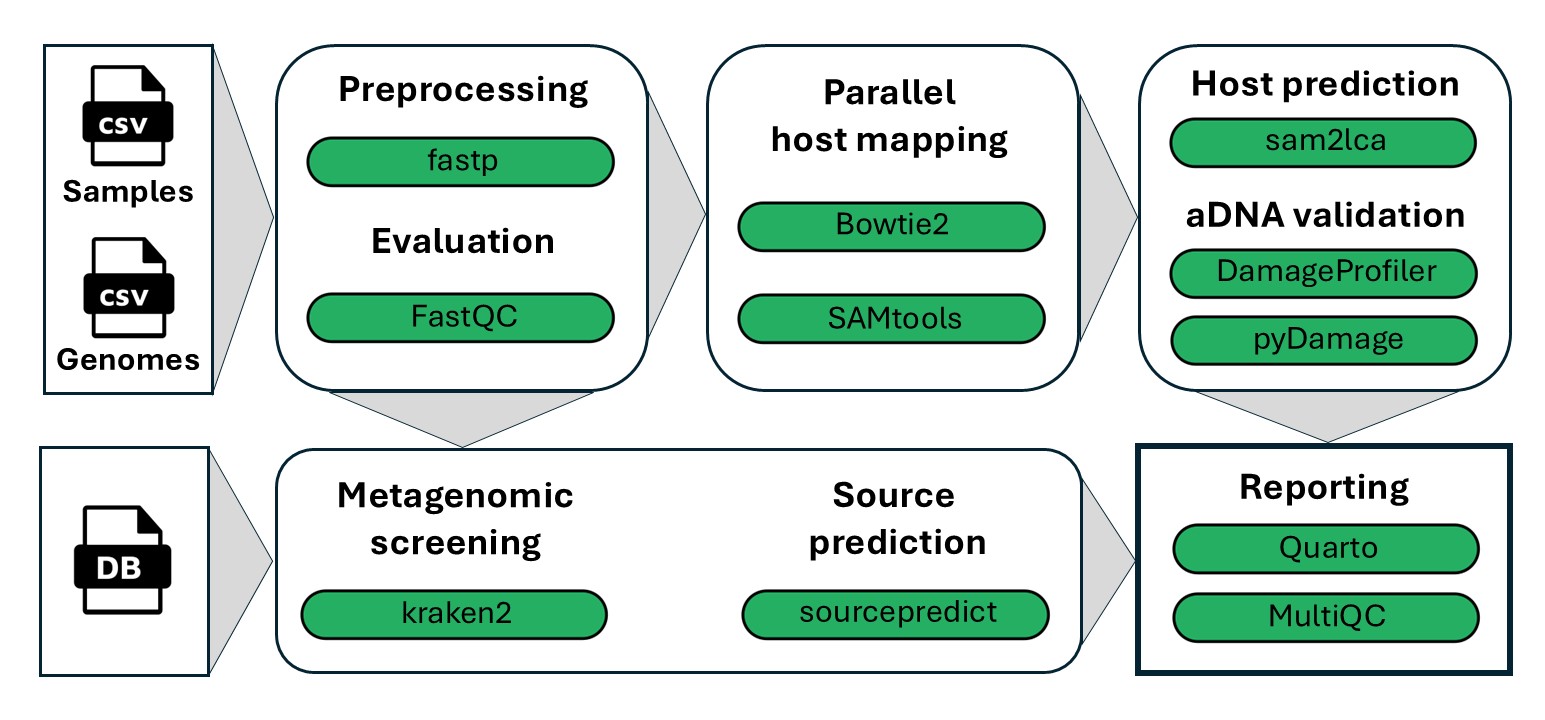
Wokflow overview
- Read QC (
FastQC) - Fastp to remove adapters and low-complexity reads (
fastp) - Mapping or reads to multiple reference genomes (
Bowtie2) - Lowest Common Ancestor analysis to retain only genome specific reads (
sam2lca) - Ancient DNA damage estimation with pyDamage and DamageProfiler
- Taxonomic profiling of unmapped reads (
kraken2) - Source predicting based on taxonic profiles (
sourcepredict) - Combining host and microbial predictions to calculate overall proportions.
- (
MultiQC) aggregate results of several individual modules. - ([Quartonotebook])(https://quarto.org/) creates a report with sample results.
The coproID pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies. Where possible, these processes have been submitted to and installed from nf-core/modules in order to make them available to all nf-core pipelines, and to everyone within the Nextflow community!
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
Pipeline usage First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
sample,fastq_1,fastq_2
PAIRED_END,PAIRED_END_S1_L002_R1_001.fastq.gz,PAIRED_END_S1_L002_R2_001.fastq.gz
SINGLE_END,SINGLE_END_S4_L003_R1_001.fastq.gz,Each row represents a fastq file (single-end) or a pair of fastq files (paired end).
Make sure that your reference genomes are from ncbi, so sam2lca can extract the taxid!
Second, prepare a genomesheet with your input genome references that looks as follows:
genomesheet.csv:
genome_name,taxid,genome_size,igenome,fasta,index
Escherichia_coli,562,5000000,,https://github.com/nf-core/test-datasets/raw/coproid/genomes/ecoli/genome.fa,
Bacillus_subtilis,1423,4200000,,https://github.com/nf-core/test-datasets/raw/coproid/genomes/bsubtilis/genome.fa,Before running the pipeline, you need to download a kraken2 database, and supply this to the pipeline using —kraken2_db The kraken2 database can be a directory or *.tar.gz
You also need to create/download the reference files for sourcepredict. These include the source anf label files, for more information see sourcepredict
Now, you can run the pipeline using:
nextflow run nf-core/coproid \
-profile <docker/singularity/.../institute> \
--input samplesheet.csv \
--genome_sheet genomesheet.csv \
--kraken2_db 'PATH/TO/KRAKENDB' \
--sp_sources 'PATH/TO/SOURCEPREDICT/SOURCES/FILE' \
--sp_labels 'PATH/TO/SOURCEPREDICT/LABELS/FILE' \
--outdir <OUTDIR>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters;
see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/coproid was originally written by Maxime Borry & Meriam Van Os.
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #coproid channel (you can join with this invite).
Citations
If you use nf-core/coproid for your analysis, please cite it using the following doi: 10.5281/zenodo.7292889
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.